|
Zhaoze Wang
I'm a PhD student at the GRASP (General Robotics, Automation, Sensing & Perception)
Lab and the Computational Neuroscience Initiative (CNI) at the University of Pennsylvania.
Email / Google Scholar / GitHub / LinkedIn / X |

|
Research InterestsI build spatial world models for embodied AI. My research focuses on how agents can learn compact spatial representations from multi-sensory inputs to support navigation, planning, and manipulation. My recent work develops RNN-based world models that enable goal-directed path planning in visually realistic environments (Habitat-Sim), with applications to robot navigation and embodied agents [REMI @ NeurIPS 2025]. I also build tools for the community: BtnkMAE (compact vision encoder), RatatouGym (navigation simulation), and OctoVLA (vision-language-action models for manipulation). My research explores how the brain builds spatial representations to support memory, navigation, and planning. I study how hippocampal place cells and grid cells emerge from learning temporally continuous sensory experiences [Time Makes Space, Trading Place for Space], and how these representations support cue-triggered goal retrieval and mental simulation of planned routes [REMI]. My most recent work proposes a unified, system-level theory of the hippocampus-MEC loop, accepted to NeurIPS 2025. |
Publications |
|

|
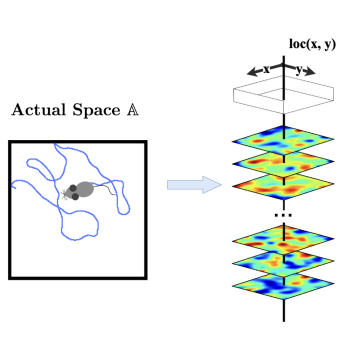
REMI: Reconstructing Episodic Memory During Internally Driven Path Planning
NeurIPS, 2025 Zhaoze Wang, Genela Morris†, Dori Derdikman†, Pratik Chaudhari†, Vijay Balasubramanian† Paper / Project Page / Simulation Suite / Vision Encoder We propose REMI, a unified, system-level theory of the hippocampus-medial entorhinal cortex (MEC) loop. The model explains how interactions between known spatial representation cell types could support cue-triggered goal retrieval, path planning, and reconstruction of sensory experiences along planned routes. An RNN-based spatial world model enabling cue-triggered path planning in visually realistic environments (Habitat-Sim). Integrates a ViT-based vision encoder (BtnkMAE) with a compact recurrent planning module for goal-directed navigation from visual inputs. Trained with multi-GPU DDP and mixed precision for scalable simulation experiments. |
|
Time Makes Space: Emergence of Place Fields in Networks Encoding Temporally Continuous Sensory Experiences
NeurIPS, 2024 Zhaoze Wang†, Ronald W. Di Tullio†, Spencer Rooke, Vijay Balasubramanian Paper / Code / Video / Project Page We model hippocampal CA3 as an autoencoder trained to recall temporally continuous sensory experiences during spatial traversal, where place cell like activity emerges in the hidden layer. The model reproduces key CA3 properties, including remapping, orthogonal representations, and stability during continual learning. See also Trading Place for Space. Demonstrates emergence of localization-relevant spatial representations from unsupervised learning on continuous sensory streams. An RNN trained to autoencode sensory signals develops spatially localized representations that reorganize across environments and recover with relearning, forming a scaffold for continual multimodal sensory integration. See also Trading Place for Space. |
|
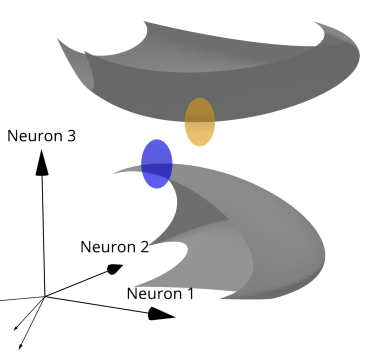
Trading Place for Space: Increasing Location Resolution Reduces Contextual Capacity in Hippocampal Codes
NeurIPS 🏆 Oral Presentation, 2024 Spencer Rooke, Zhaoze Wang, Ronald W. Di Tullio, Vijay Balasubramanian Paper / Video We showed that increasing the resolution of spatial encoding reduces the number of distinct contexts that can be stored by place cells, revealing a trade-off between position accuracy and contextual capacity. We derived theoretical bounds on this trade-off using manifold geometry and neural noise models. See also Time Makes Space. A geometric framework quantifying trade-offs between spatial resolution and contextual capacity in self-localizing systems. Modeling population activity as high-dimensional manifolds, we show that separable contexts scale exponentially with network size but trade off with spatial resolution. See also Time Makes Space. |
Opensource |
|

|
OctoVLA: Vision-Language-Action Model for Manipulation
Source Code Deployed Octo, a generalist VLA model, on the LIBERO manipulation benchmark with language-conditioned control. Evaluated model performance across 10 manipulation tasks with varying language instructions. Integrating BtnkMAE as a compact vision encoder to compare sample efficiency with the default ViT encoder. |
|
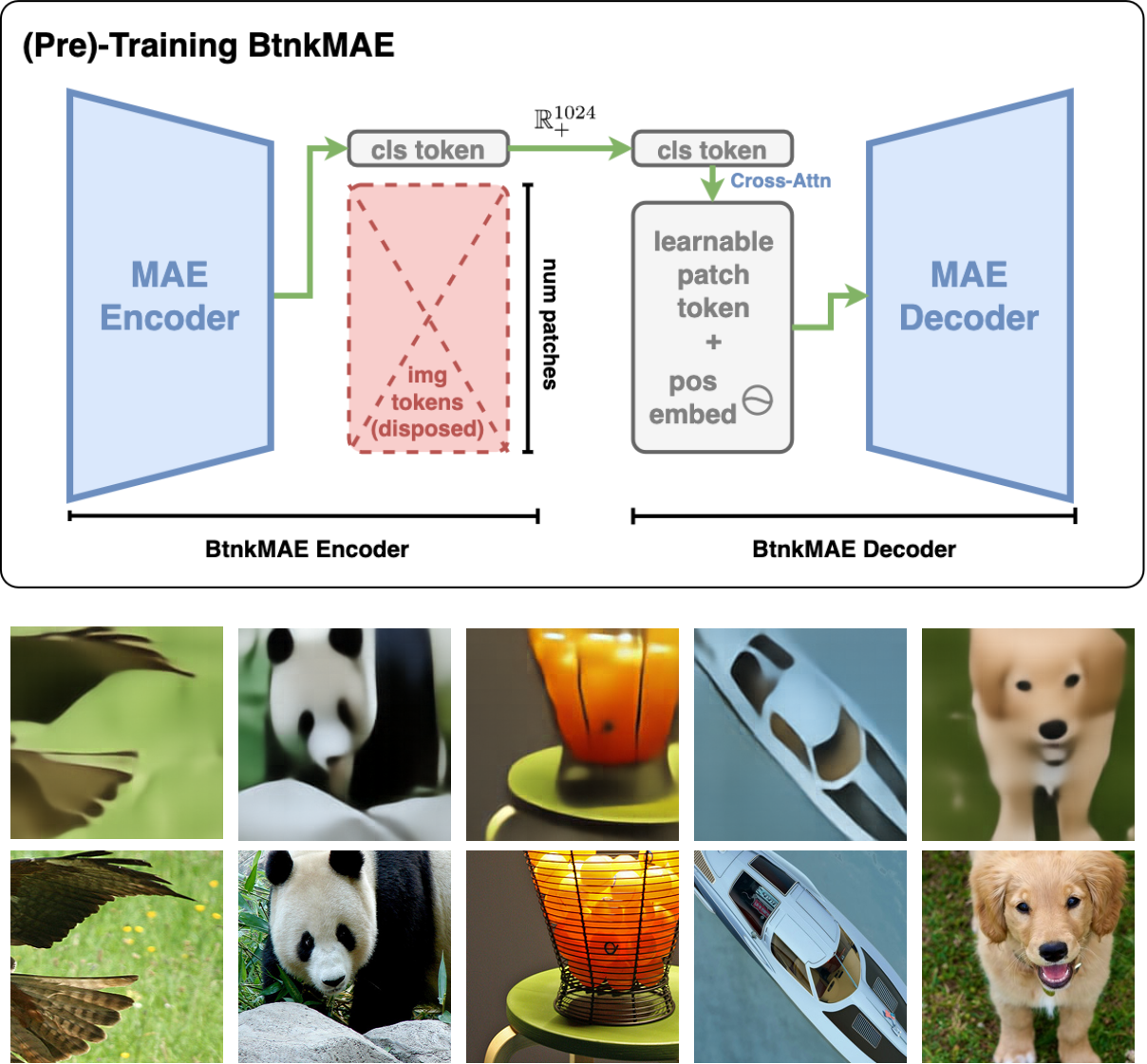
BtnkMAE: Compact Vision Encoder for Embodied AI
Source Code Compact vision encoder compressing images to 1024-D decodable embeddings, 196× more compact than standard ViT. Pretrained ViT-B/L on ImageNet-1K using DDP across 6×RTX 4090 GPUs. Designed for embodied AI applications where memory and bandwidth are constrained. |
|
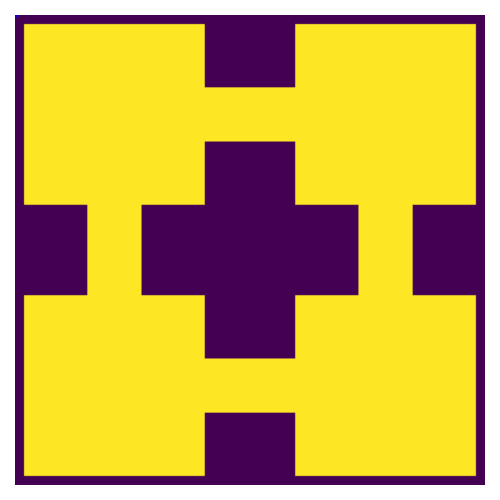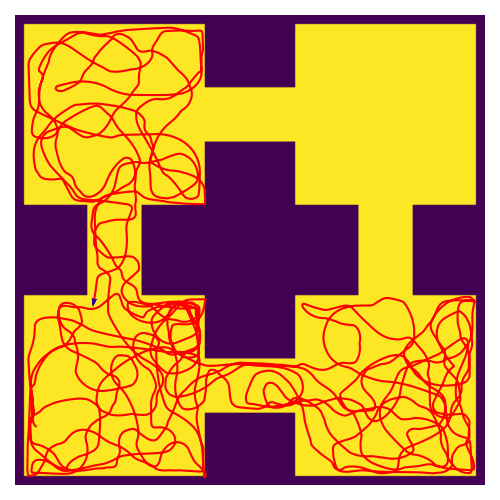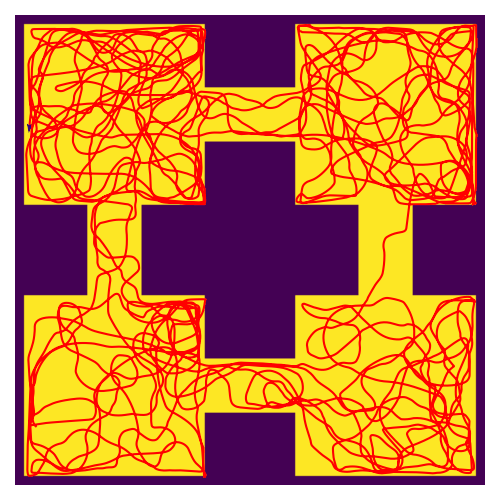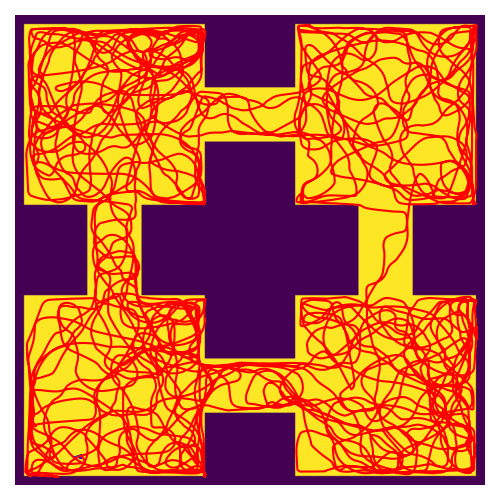
RatatouGym: Navigation Simulation Framework
Documentation / Source Code Open-source navigation framework that directly simulates sensory inputs during navigation. Engineered modular backends (FAISS, K-d tree, PyTorch) for scalable multi-agent experiments. Used in REMI (NeurIPS 2025) for training spatial world models. |
|
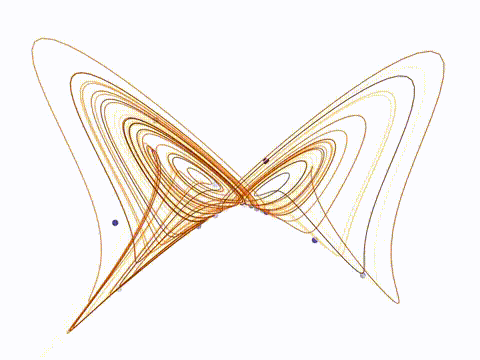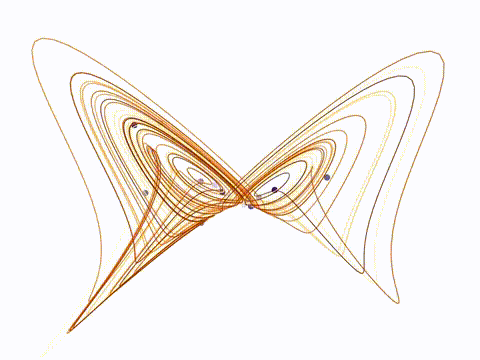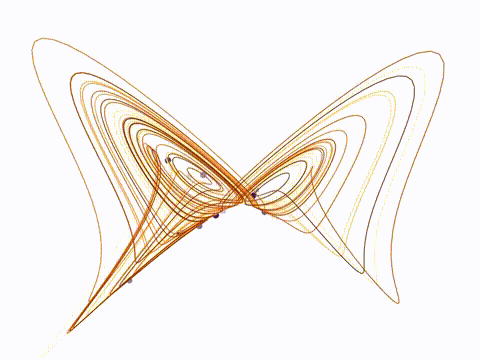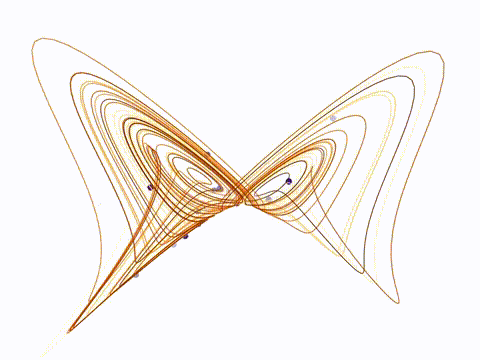
NN4N: Modular RNN Framework
Documentation / Source Code PyTorch framework for hierarchical RNNs with custom autograd, connectivity constraints, and optimizers. Supports biologically plausible architectures with sparse, structured connectivity. Published on PyPI with 6K+ downloads. |
Miscellanea |
|
Service
|
Teaching Assistant, ESE 5460: Principles of Deep Learning, Fall 2025 Teaching Assistant, PHYS 5585: Comp. and Theoretical Neurosci., Spring 2026 |
|
Design and code of this website is adapted from Jon Barron's website. |


